Current Research
Secondary (ion-gradient driven) active transport proteins represent a core group of functional processes which are crucial in both microbial adaptations and human physiology and disease. Although transport protein families tend to display high evolutionary conservation in sequence and overall structure, they also display high functional variations between homologs, implying that relatively few side chain changes may account for key local effects on the active-site conformation and function.
We are interested to gain insight on side chains and sequence motifs that dictate substrate specificity and uptake in such transporter families by using bacterial homologs as study models. We currently focus on the evolutionarily broad family of nucleobase-ascorbate transporters (NAT/NCS2) which includes proteins responsible for the uptake of several frontline purine-related drugs or analogues.
A major challenge for the study of NAT/NCS2 family is that very few of the many thousand predicted members have been identified functionally, albeit indicating high heterogeneity in specificities, whereas high-resolution structures or models thereof have been developed only recently. Research on this family has drawn considerable attention with the elucidation of x-ray structures of two homologs, the uracil permease UraA of E. coli (Lu et al., 2011; Yu et al., 2017) and the xanthine/uric acid permease UapA of Aspergillus nidulans (Alguel et al., 2016), coupled with mutagenesis studies on UapA (Diallinas, 2016;Kourkoulou et al., 2018) and the xanthine permease XanQ from our lab (Frillingos, 2012; Karena et al., 2015). These transporters are suggested to function as homodimers, following an elevator-type mechanism (Alguel et al., 2016; Yu et al., 2017). They also are structural and mechanistic prototypes for a group of transporter families in the APC superfamily including SLC4, SLC23 and SLC26 (Chang and Geertsma, 2017).
We are focusing mostly on bacterial members of the NAT/NCS2 family which is highly expanded and functionally diversified in certain bacterial taxa, including primarily metabolically versatile species from proteobacteria, actinobacteria and firmicutes (Chaliotis et al., 2018). To understand the molecular basis of substrate-specificity divergence in NAT/NCS2 family, we follow three major experimental routes:
· (1) Systematic structure-function analysis of a representative homolog, the xanthine permease XanQ from E. coli K-12, cloned and characterized in our lab. This approach has been based on Cys-scanning and site-directed alkylation analysis and yielded a comprehensive view of the functionally important residues in XanQ (Karena et al., 2015).
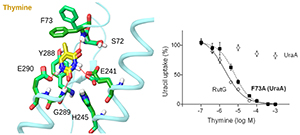
· (2) Functional and mutational characterization of a series of NAT/NCS2 homologs from various bacteria to reveal correlation patterns between residues present at important positions and substrate specificity profiles. Such data offer a basis to understand mechanisms of substrate-profile divergence between different homologs and design efficient strategies to modulate specificity to novel directions. In the context of this line of studies, we have provided novel insight on the basis of specificity divergence in the NAT/NCS2 cluster of uracil and uracil/thymine transporters (Botou et al., 2018).
· (3) Reconstruction and analysis of ancestral homologs and cross-homolog chimeras between present-day NAT/NCS2 homologs with different specificities, to elucidate structural elements and interactions which allow acquisition of new functions and substrate-recognition profiles.
Our work on the NAT/NCS2 family has included development of specific site-directed alkylation protocols (Fig. 1), formulation of structure-functional models (Fig. 2), and collaborations for molecular dynamics simulations and bioinformatics studies (Fig. 3).
Apart from mechanistic and evolutionary implications, our research confers to elucidation of the role of NAT/NCS2 transporters to distinct bacterial adaptations (Botou et al., 2020;Botou et al., 2018) and is expected also to provide insight on the nucleobase/nucleoside transport systems of commensal and pathogenic bacteria of the human gut.